RNA interference
 RNAi is a mechanism of genome regulation conserved in nearly all eukaryotes. It is mediated by small RNAs, which associate with Argonaute effector proteins to direct sequence-specific regulation. How this core mechanism is deployed is variable within and between species: RNAi pathways can target genes, transposons, or other repetitive sequences, and can exert repression of transcription, stability, or translation of target RNAs. Our lab seeks to elucidate fundamental mechanisms of RNAi including the factors that determine different regulatory functions, as well as understand the biological impact of RNAi on genome regulation and stability, with a focus on paradigmatic examples in fungi.
RNAi is a mechanism of genome regulation conserved in nearly all eukaryotes. It is mediated by small RNAs, which associate with Argonaute effector proteins to direct sequence-specific regulation. How this core mechanism is deployed is variable within and between species: RNAi pathways can target genes, transposons, or other repetitive sequences, and can exert repression of transcription, stability, or translation of target RNAs. Our lab seeks to elucidate fundamental mechanisms of RNAi including the factors that determine different regulatory functions, as well as understand the biological impact of RNAi on genome regulation and stability, with a focus on paradigmatic examples in fungi.
RNAi-mediated genome regulation in Cryptococcus
 Cryptococcus is an opportunistic human fungal pathogen that kills over one hundred thousand people annually. Treatment options are limited, and emergence of resistance to existing antifungal drugs is a growing problem. A primary role of RNAi in Cryptococcus appears to be the silencing of transposable elements, and loss of RNAi leads to increased rates of transposon-mediated mutation that can potentiate increased virulence or drug resistance. However, how RNAi operates in Cryptococcus, a member of the little-studied basidiomycete branch of the fungal kingdom, remains unclear. We have recently initiated a major new programme of research that aims to elucidate novel aspects of RNAi function and regulation in this clinically important pathogen, and understand how modulation of RNAi may contribute to fungal adaptation.
Cryptococcus is an opportunistic human fungal pathogen that kills over one hundred thousand people annually. Treatment options are limited, and emergence of resistance to existing antifungal drugs is a growing problem. A primary role of RNAi in Cryptococcus appears to be the silencing of transposable elements, and loss of RNAi leads to increased rates of transposon-mediated mutation that can potentiate increased virulence or drug resistance. However, how RNAi operates in Cryptococcus, a member of the little-studied basidiomycete branch of the fungal kingdom, remains unclear. We have recently initiated a major new programme of research that aims to elucidate novel aspects of RNAi function and regulation in this clinically important pathogen, and understand how modulation of RNAi may contribute to fungal adaptation.
RNAi-directed chromatin modification in fission yeast
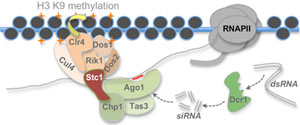 In the fission yeast Schizosaccharomyces pombe, RNAi functions primarily to promote assembly of heterochromatin at centromeres through the targeting of repetitive pericentromeric transcripts. This system has therefore become a valuable model for studying RNAi-directed chromatin modification and transcriptional silencing. Our lab has a longstanding interest in elucidating novel factors and mechanisms that contribute to RNAi-directed chromatin modification in S. pombe, in particular dissecting the interplay between RNAi and other epigenetic regulators in promoting and regulating the initiation and propagation of heterochromatic domains. We are also interested in understanding the evolution of specialisation in RNAi function through comparative analyses across the fission yeast clade.
In the fission yeast Schizosaccharomyces pombe, RNAi functions primarily to promote assembly of heterochromatin at centromeres through the targeting of repetitive pericentromeric transcripts. This system has therefore become a valuable model for studying RNAi-directed chromatin modification and transcriptional silencing. Our lab has a longstanding interest in elucidating novel factors and mechanisms that contribute to RNAi-directed chromatin modification in S. pombe, in particular dissecting the interplay between RNAi and other epigenetic regulators in promoting and regulating the initiation and propagation of heterochromatic domains. We are also interested in understanding the evolution of specialisation in RNAi function through comparative analyses across the fission yeast clade.


